Morphologica Save
A library of supporting code for numerical modelling (JSON config, HDF5 data, Modern OpenGL visualization)
morphologica

Header-only library code to visualize C++ numerical simulations using fast, modern OpenGL.
Morphologica can run standalone (using GLFW for window handling) and it is also Qt and wxWidgets compatible!
NEW: Morphologica can now be used with wxWidgets! For example code, see examples/wx/
NEW: Morphologica now has OpenGL Compute Shader managers! See examples/gl_compute for details.
NEW: Now morphologica is compatible with the Raspberry Pi! See examples/pi/. These examples show how to set the OpenGL version to 3.1 ES, which is Pi compatible.
You'll find all of the library code in the morph directory and you can find example code and screenshots here.
morphologica has an (in-progress) documentation and reference website at https://abrg-models.github.io/morphologica/.
Quick Start
This quick start shows dependency installation for Linux, because on this platform, it's a single call to apt (or your favourite package manager). If you're using a Mac, see README.build.mac for help getting dependencies in place. It's README.build.windows for Windows users.
# Install dependencies for building graph1.cpp and (almost) all the other examples (assuming Debian-like OS)
sudo apt install build-essential cmake git wget \
freeglut3-dev libglu1-mesa-dev libxmu-dev libxi-dev \
libglfw3-dev libfreetype-dev libarmadillo-dev libhdf5-dev
git clone [email protected]:ABRG-Models/morphologica # Get your copy of the morphologica code
cd morphologica
mkdir build # Create a build directory
cd build
cmake .. # Call cmake to generate the makefiles
make graph1 # Compile a single one of the examples. Add VERBOSE=1 to see the compiler commands.
./examples/graph1 # Run the program. You should see a graph of a cubic function.
# After closing the graph1 program, open its source code and modify something (see examples/graph2.cpp for ideas)
gedit ../examples/graph1.cpp
The program graph1.cpp is:
// Visualize a graph. Minimal example showing how a default graph appears
#include <morph/Visual.h>
#include <morph/GraphVisual.h>
#include <morph/vvec.h>
int main()
{
// Set up a morph::Visual 'scene environment'.
morph::Visual v(1024, 768, "Made with morph::GraphVisual");
// Create a new GraphVisual object with offset within the scene of 0,0,0
auto gv = std::make_unique<morph::GraphVisual<double>> (morph::vec<float>({0,0,0}));
// Boilerplate bindmodel function call - do this for every model you add to a Visual
v.bindmodel (gv);
// Data for the x axis. A vvec is like std::vector, but with built-in maths methods
morph::vvec<double> x;
// This works like numpy's linspace() (the 3 args are "start", "end" and "num"):
x.linspace (-0.5, 0.8, 14);
// Set a graph up of y = x^3
gv->setdata (x, x.pow(3));
// finalize() makes the GraphVisual compute the vertices of the OpenGL model
gv->finalize();
// Add the GraphVisual OpenGL model to the Visual scene (which takes ownership of the unique_ptr)
v.addVisualModel (gv);
// Render the scene on the screen until user quits with 'Ctrl-q'
v.keepOpen();
return 0;
}
The program generates a clean looking graph...
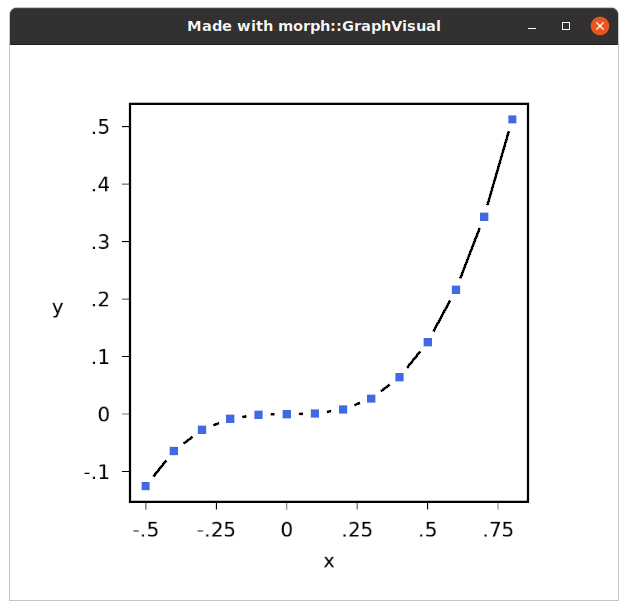
...and the code compares favourably (in terms of amount of boilerplate code) with the equivalent Python, graph1.py:
# Visualize the graph from graph1.cpp in Python
import matplotlib.pyplot as plt
import numpy as np
# Create some data for the x axis
x = np.linspace(-0.5, 0.8, 14)
# Set a graph up of y = x^3
plt.plot(x, np.power(x,3), '-o')
# Add labels
plt.title('Made with Python/numpy/matplotlib')
plt.xlabel('x')
plt.ylabel('y')
# Render the graph on the screen until user quits with 'q'
plt.show()
See the coding README for a description of some of the main classes that morphologica provides.
What is morphologica?
This header-only C++ code provides simulation support facilities for simulations of dynamical systems, agent-based models or, in fact, any program that needs dynamic, runtime visualization.
It helps with:
-
Visualizing your model while it runs. A modern OpenGL visualization scheme called morph::Visual provides the ability to visualise 2D and 3D graphs of surfaces, lines, bars, scatter plots and quiver plots with minimal processing overhead. Here's a morph::Visual helloworld and a more complete example. It's almost as easy to draw a graph in C++ with morphologica as it is to do so in Python.
-
Configuration: morphologica allows you to easily set up a simulation parameter configuration system, using the JSON reading and writing abilities of morph::Config. (morph::Config Example)
-
Saving data from your simulation. morphologica provides a set of easy-to-use convenience wrappers (morph::HdfData) around the HDF5 C API. By saving data in a standard format, it is easy to access simulation data in python, MATLAB or Octave for analysis and graphing. (HdfData Example)
It keeps out of the way of what kind of simulation you write. Our programs typically start with some kind of preamble, in which we use morph::Config to load up a JSON parameter file defining the values of the parameters for the simulation run. We might also use morph::HdfData to retrieve some data (e.g. the state) from an earlier simulation and then set up a morph::Visual object for the visualization. We then might call a function, or create a class object which defines the simulation. This may or may not access features from the morphologica headers.
As the simulation progresses, we update the data in the morph::Visual scene; save images from the scene for movie making and record data as often as we want it using morph::HdfData. At the end of the program, as well as saving any final data, we use morph::Config to save out a 'version of record' of the parameters that were used, along with git information which morph::Config can extract so that we could find the exact version of the simulation for future reproduction of the result.
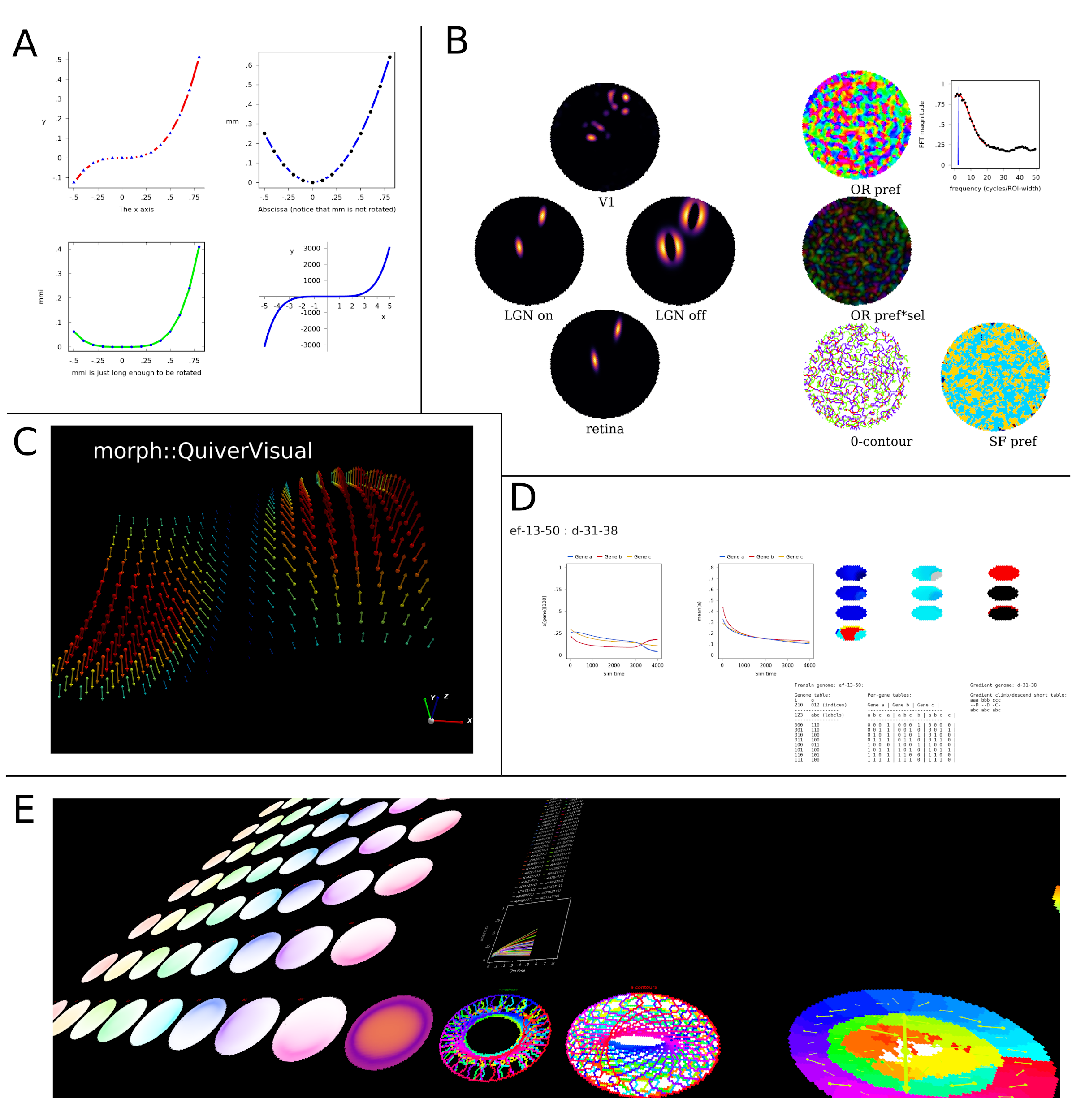
A selection of visualisations made with morphologica. A 2D graphs. B A self-organising map simulation (orientation preference maps). C Three dimensional quiver plot. D gene driven reaction diffusion model. E Debugging a large model.
Although it need not be incorporated into your actual simulation, morphologica does also provide classes that you might find useful. Examples include:
-
morph::HexGrid and morph::CartGrid: classes for running simulations on hexagonal or Cartesian grids (managing all the neighbour relationships between elements and allowing you to specific various boundary shapes for your domain). See also morph::ColourBarVisual to draw colour bars for your morph::ColourMap of choice.
-
morph::vec and morph::vvec: Cool mathematical vector classes - these are like std::vector and std::array but they also sport maths methods. vvec usage example.
-
morph::MathAlgo a class containing mathematical algorithms.
-
morph::BezCurve and friends: classes for working with Bezier curves.
-
morph::Winder A class to compute the winding number of a path.
-
morph::Scale A class for simple scaling/transformation of numbers.
-
morph::NM_Simplex and morph::Anneal Optimization algorithms. Example simulated annealing usage and the Nelder-Mead simplex method
-
morph::RandUniform and friends. Wrapper classes around C++'s high quality random number generation code (Usage example).
-
morph::ReadCurves Code to read SVG drawings to turn Bezier-curve based lines into paths containing evenly spaced coordinates.
morphologica is a way of storing our 'group knowledge' for posterity.
Some existing projects which use morphologica are:
- BarrelEmerge A reaction-diffusion style model: https://github.com/ABRG-Models/BarrelEmerge
- RetinoTectal Reaction-diffusion and agent-based modelling: https://github.com/sebjameswml/RetinoTectal
- ArtificialGeneNets Neural networks: https://github.com/stuartwilson/ArtificialGeneNets
Code documentation
See README.coding.md for a guide to the main classes.
morphologica code is enclosed in the morph namespace. If README.coding.md doesn't coverit, then the header files (They're all in morph/) contain code documentation.
You can find example programs which are compiled when you do the standard cmake-driven build of morphologica in both the tests/ subdirectory and the examples/ subdirectory. The readme in examples/ has some nice screenshots.
For full, compilable, standalone examples of the code, see the standalone_examples/ subdirectory. You can use these as templates for creating your own projects that use morphologica library code.
For more info on how to set up CMake files to build a program using morphologica (and some hints as to what you'll need to do with an alternative directed build system), see README.cmake.md.
Credits
Authorship of morphologica code is given in each file. Copyright in the software is owned by the authors.
morphologica is made possible by a number of third party projects whose source code is included in this repository. These include nlohmann::json, lodepng, rapidxml and incbin. Thanks to the authors of these projects!
morphologica is distributed under the terms of the Apache License, version 2 (see LICENSE.txt).
