MinSizeKmeans Save
A python implementation of KMeans clustering with minimum cluster size constraint (Bradley et al., 2000)
MinSizeKmeans
A python implementation of KMeans clustering with minimum cluster size constraint (Bradley et al., 2000)[1]
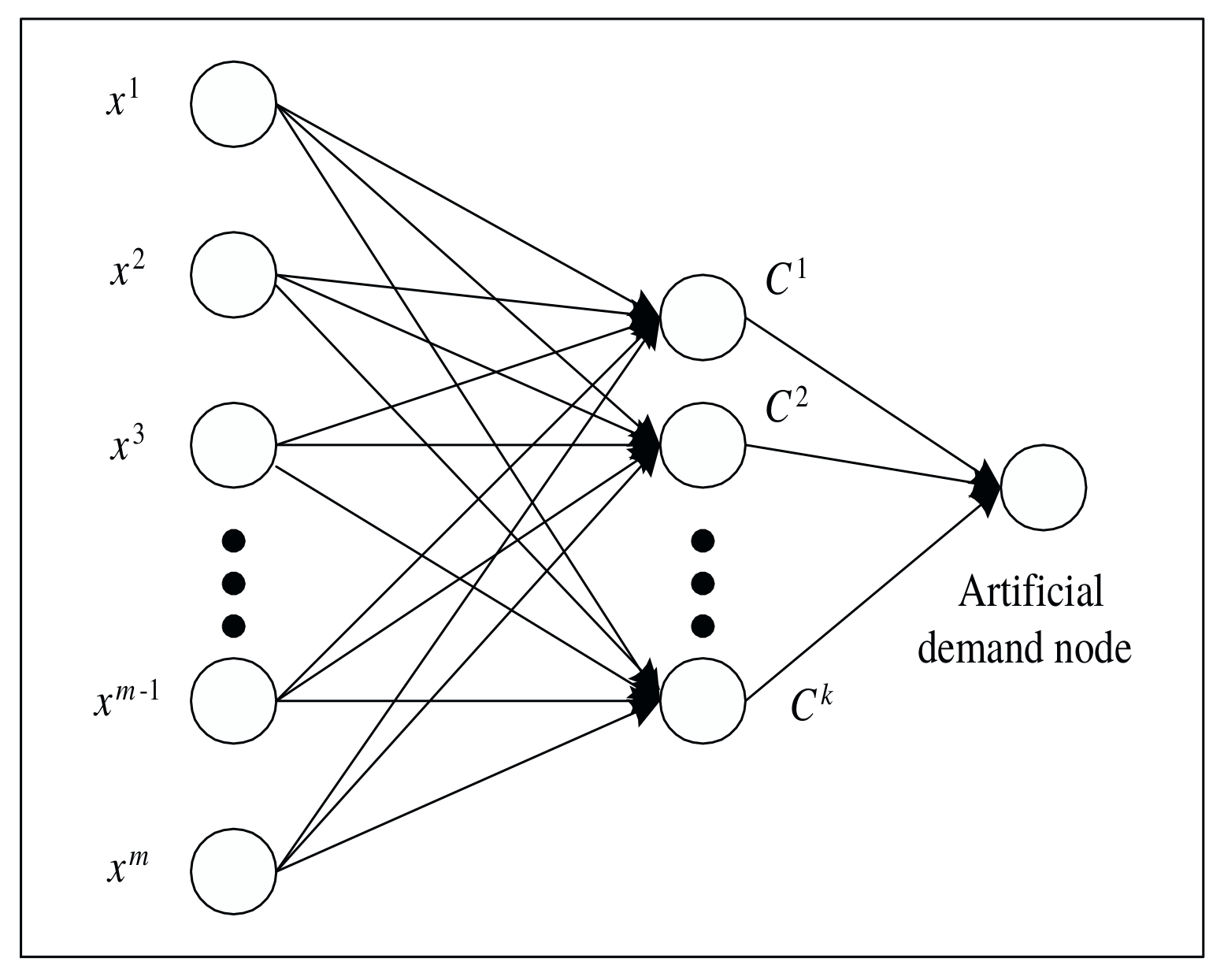
Note that the paper suggests using a dedicated minimum-cost network flow algorithm for solving the subproblem, but in my implementation I use standard MIP solvers. My motivation for doing so was this comment from Tobias Achterberg:
If you have a pure network, then the constraint matrix is totally unimodular, so any vertex solution will be integral. And since the simplex algorithm always returns a vertex solution (if one exists), your solution will be integral.
Note that in our experience, a network simplex algorithm is not faster than the dual simplex, which is the reason why Gurobi does not provide a network simplex.
Usage
usage: run_mskmeans.py [-h] [-n NUM_ITER] [-o OUTFILE] datafile k min_size
positional arguments:
datafile file containing the coordinates of instances
k number of clusters
min_size minimum size of each cluster
optional arguments:
-h, --help show this help message and exit
-n NUM_ITER, --NUM_ITER NUM_ITER
run the algorithm for NUM_ITER times and return the best clustering
-o OUTFILE, --OUTFILE OUTFILE
store the result in OUTFILE
To see a run of algorithm on example data, run the script example_run.sh in minsize_kmeans directory.
Dependencies
This program uses Pulp to solve the assignment subproblem. You should also have a MIP solver (Gurobi, Cplex, Cbc, etc.) installed and have it configured to work with Pulp.
Extension
This algorithm can be easily extended to account for a constraint on maximum number of instances in a cluster. To do so, in the network flow formulation of [1], we add constraints on the capacity of arcs that enter the sink node.
To run the algorithm with both minimum and maximum size constraints, use this command:
usage: minmax_kmeans.py [-h] [-n NUM_ITER] [-o OUTFILE] datafile k min_size max_size
positional arguments:
datafile file containing the coordinates of instances
k number of clusters
min_size minimum size of each cluster
max_size maximum size of each cluster
optional arguments:
-h, --help show this help message and exit
-n NUM_ITER, --NUM_ITER NUM_ITER
run the algorithm for NUM_ITER times and return the best clustering
-o OUTFILE, --OUTFILE OUTFILE
store the result in OUTFILE
An example run is included in the script example_run.sh in minsize_kmeans directory.
Constrained K-Means with Weighted Instances
Sometimes instead of the size of clusters, we want to constrain the total weight of instances that are in each cluster. Unfortunately, the subproblem for constrained k-means in this case will be NP-complete. However, we can still give this subproblem to a MIP solver and hope that it will be solved in a reasonable time. To run this algorithm, use this command:
usage: weighted_mm_kmeans.py [-h] [-n NUM_ITER] [-o OUTFILE]
datafile k weightfile min_weight max_weight
positional arguments:
datafile file containing the coordinates of instances
k number of clusters
weightfile file containing the weights of instances
min_weight minimum total weight for each cluster
max_weight maximum total weigth for each cluster
optional arguments:
-h, --help show this help message and exit
-n NUM_ITER, --NUM_ITER NUM_ITER
run the algorithm for NUM_ITER times and return the best clustering
-o OUTFILE, --OUTFILE OUTFILE
store the result in OUTFILE
An example run is included in the script example_weighted.sh in minsize_kmeans directory.
References
- Bradley, P. S., K. P. Bennett, and Ayhan Demiriz. "Constrained k-means clustering." Microsoft Research, Redmond (2000): 1-8.
