Centrifuge Toolkit Save
Tool for visualizing and empirically analyzing information encoded in binary files
Centrifuge
Centrifuge makes it easy to use visualization, statistics and machine learning to analyze information in binary files.
This tool implements two new approaches to analysis of file data:
-
DBSCAN, an unsupervised machine learning algorithm, is used to find clusters of byte sequences based on their statistical properties (features). Byte sequences that encode the same data type, e.g. machine code, typically have similar properties. As a result, clusters are often representative of a specific data type. Each cluster can be extracted and analysed further.
-
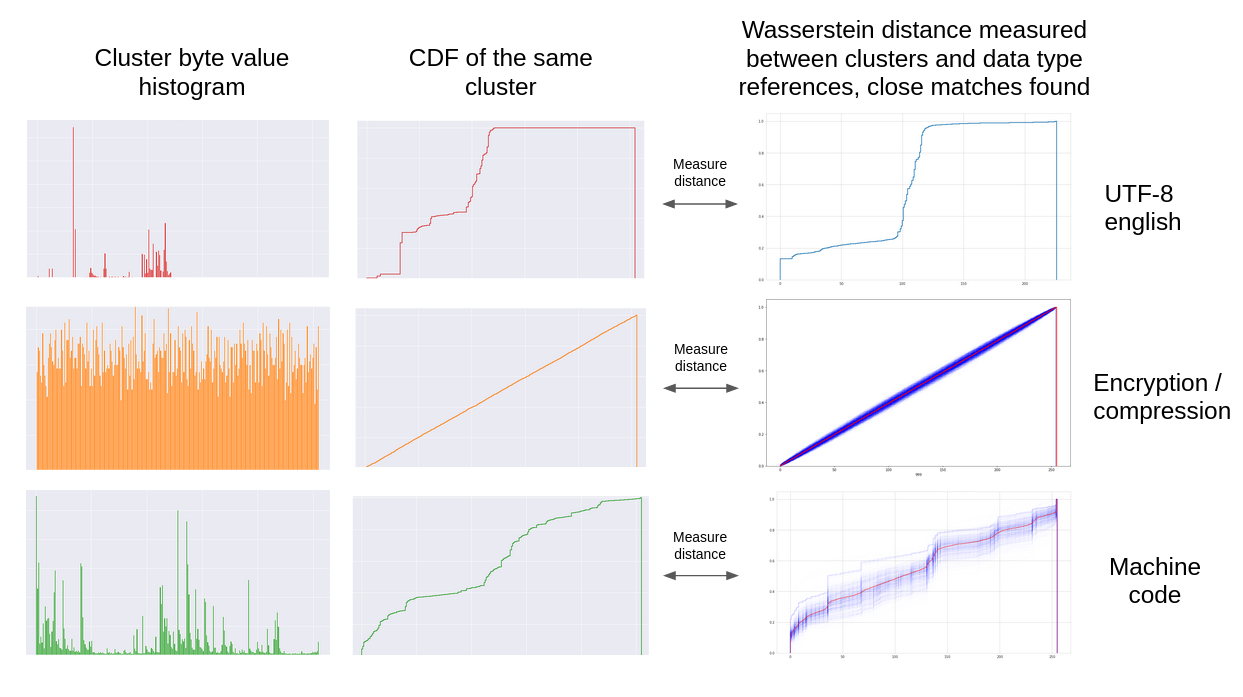
The specific data type of a cluster can often be identified without using machine learning by measuring the Wasserstein distance between its byte value distribution and a data type reference distribution. If this distance is less than a set threshold for a particular data type, that cluster will be identified as that data type. Currently, reference distributions exist for high entropy data, UTF-8 english, and machine code targeting various CPU architectures.
These two approaches are used together in sequence: first DBSCAN finds clusters, then the Wasserstein distances between the clusters' data and the reference distributions are measured to identify their data type. To identify the target CPU of any machine code discovered in the file, Centrifuge uses ISAdetect.
Required Libraries
All required libraries come bundled with Anaconda.
*Developed in a Linux environment. Not tested on Windows or MacOS.
Usage
Detailed walkthroughs can be found in the notebooks. Code snippets are located in the scripts folder.
- Introduction to Centrifuge provides an overview of Centrifuge's features and a demonstration of how the tool works.
-
Using DBSCAN to Cluster File Data shows examples of how to adjust DBSCAN's
epsandmin_samplesparameters to get the best results. - Analyzing Firmware with Centrifuge and Analyzing Firmware with Centrifuge Example 2 provide tutorials for analyzing firmware binaries.
- Analyzing Machine Code Targeting an Usupported Architecture discusses what may occur when an executable binary contains machine code targeting a CPU architecture for which there is no matching reference distribution and ISAdetect does not correctly classify it.
Overview of the Approach
The first step is file partitioning and feature measurement.
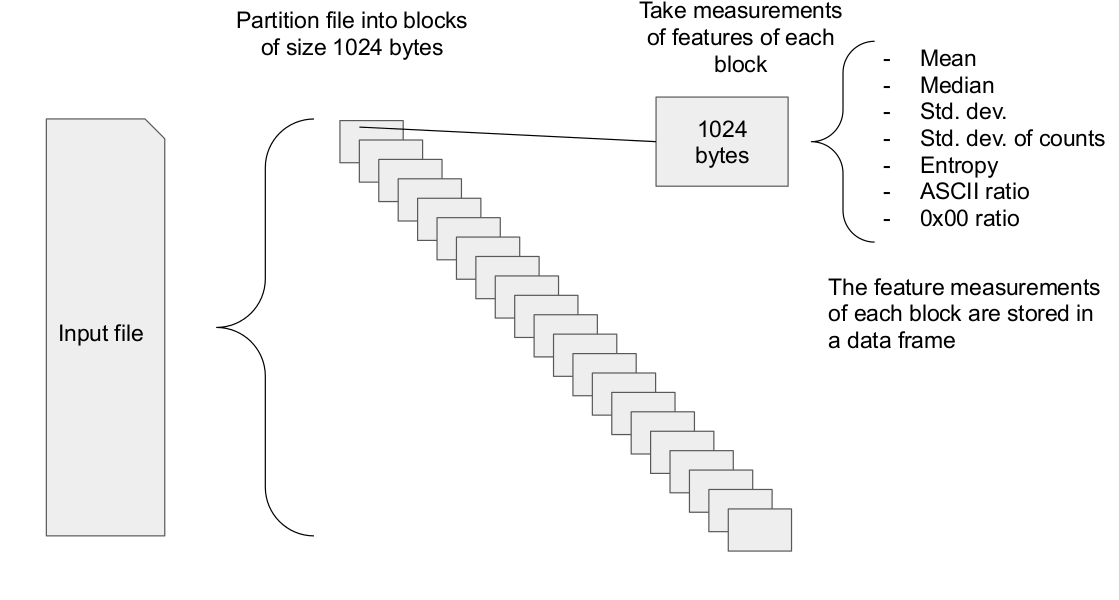
DBSCAN can then be used to find clusters in the file data.
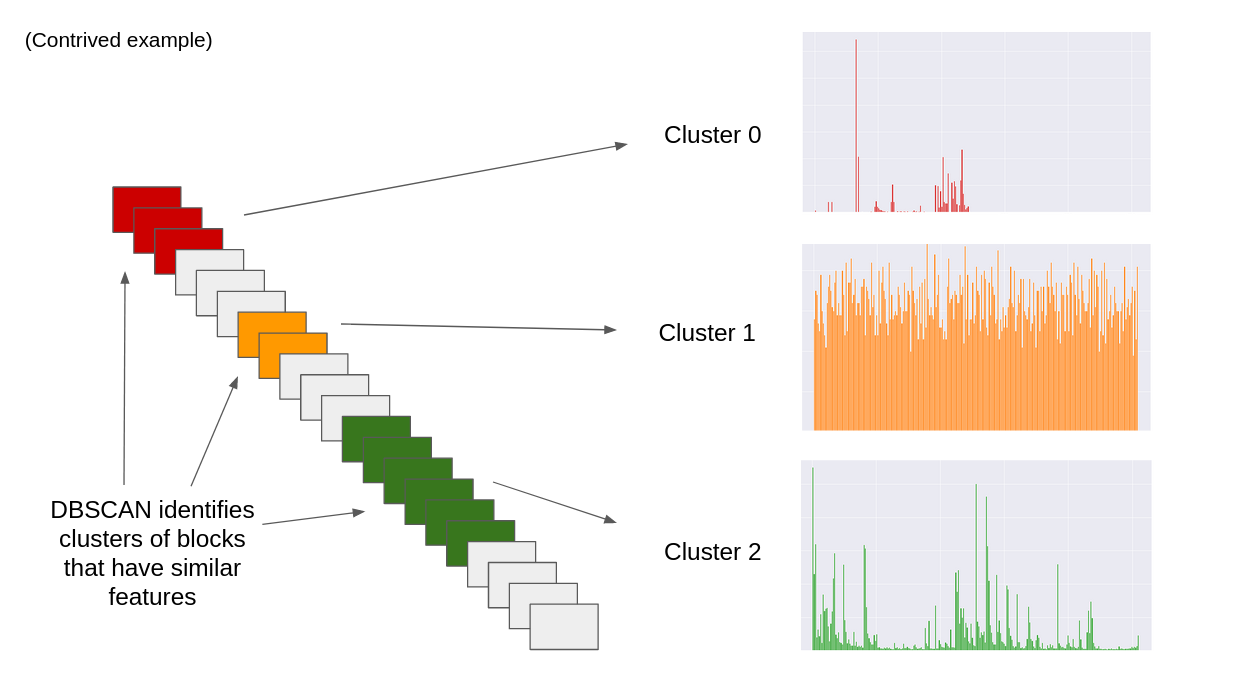
Once clusters have been found, the data in the clusters can be identified.
The feature observations of each cluster are stored in a separate data frame, one for each cluster (e.g if 6 clusters are found, there will be 6 data frames, 1 per cluster). The output of DBSCAN is also saved in a data frame. This means custom analysis of any/all clusters can easily be performed any time after DBSCAN identifies clusters in the file data.
Example Output
Output of bash.identify_cluster_data_types(), as seen in Introduction to Centrifuge:
Searching for machine code
--------------------------------------------------------------------
[+] Checking Cluster 4 for possible match
[+] Closely matching CPU architecture reference(s) found for Cluster 4
[+] Sending sample to https://isadetect.com/
[+] response:
{
"prediction": {
"architecture": "amd64",
"endianness": "little",
"wordsize": 64
},
"prediction_probability": 1.0
}
Searching for utf8-english data
-------------------------------------------------------------------
[+] UTF-8 (english) detected in Cluster 3
Wasserstein distance to reference: 16.337275669642857
[+] UTF-8 (english) detected in Cluster 5
Wasserstein distance to reference: 11.878225097656252
Searching for high entropy data
-------------------------------------------------------------------
[+] High entropy data found in Cluster 1
Wasserstein distance to reference: 0.48854199218749983
[*] This distance suggests the data in this cluster could be
a) encrypted
b) compressed via LZMA with maximum compression level
c) something else that is random or close to random.
File Data Visualization
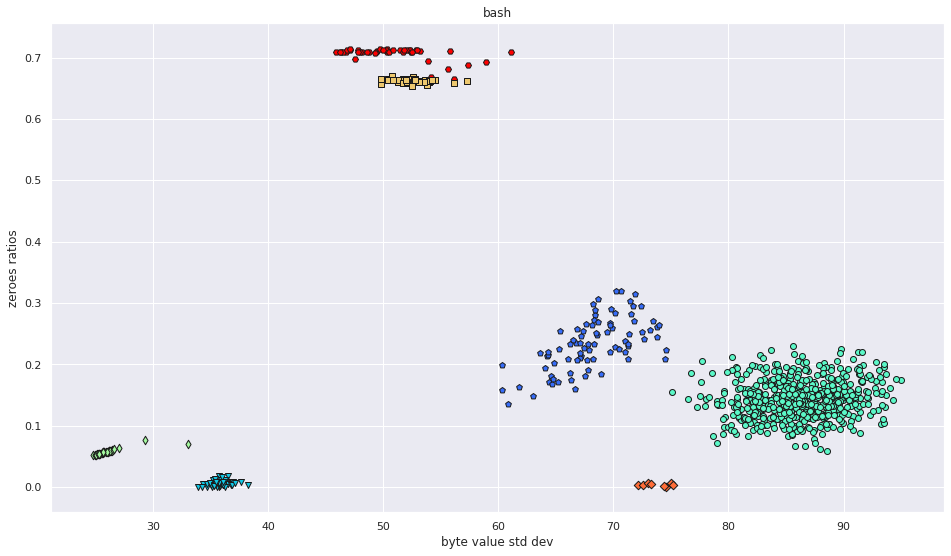
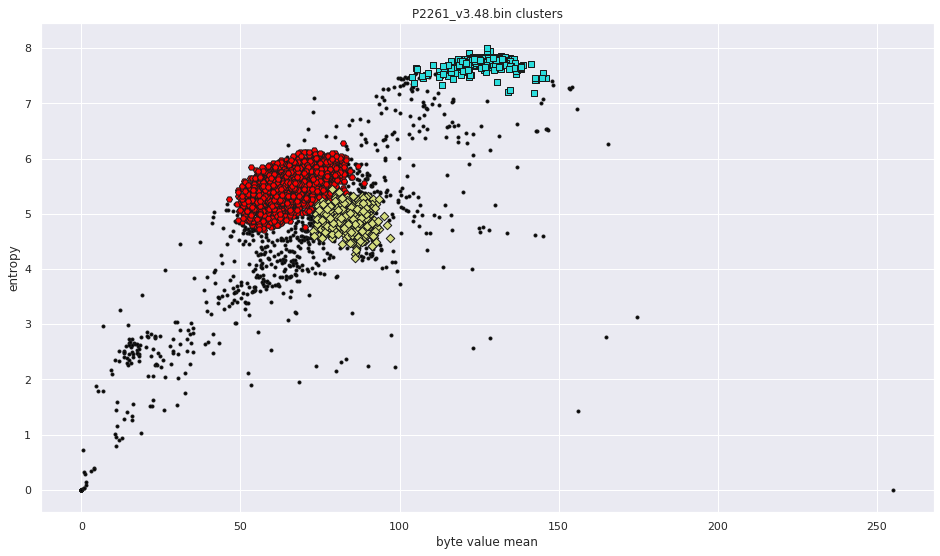
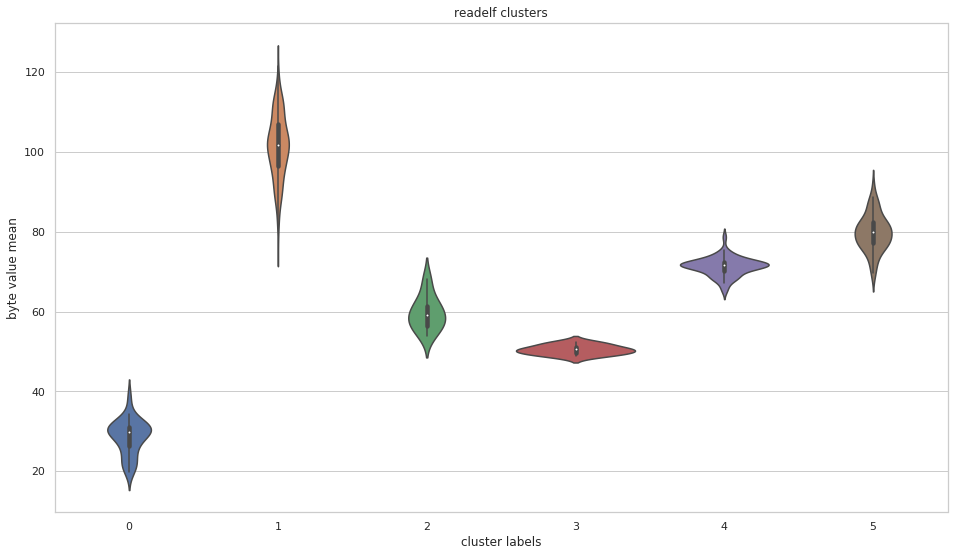
More pictures can be found in the gallery.
Example Use Cases
-
Determining whether a file contains a particular type of data.
An entropy scan is useful for discovering compressed or encrypted data, but what about other data types such as machine code, symbol tables, sections of hardcoded ASCII strings, etc? Centrifuge takes advantage of the fact that in binary files, information encoded in a particular way is stored contiguously and uses scikit-learn's implementation of DBSCAN to locate these regions.
-
Analyzing files with no metadata such as magic numbers, headers or other format information.
This includes most firmware, as well as corrupt files. Centrifuge does not depend on metadata or signatures of any kind.
-
Investigating differences between different types of data using statistical methods or machine learning, or building a model or "profile" of a specific data type.
Does machine code differ in a systematic way from other types of information encoded in binary files? Can compressed data be distinguished from encrypted data? These questions can be investigated in an empirical way using Centrifuge.
-
Visualizing information in files using Python libraries such as Seaborn, Matplotlib and Altair
Rather than generate elaborate 2D or 3D visual representations of file contents using space-filling curves or cylindrical coordinate systems, Centrifuge creates data frames that contain the feature measurements of each cluster. The information in these data frames can be easily visualized with boxplots, violin plots, pairplots, histograms, density plots, scatterplots, barplots, cumulative distribution function (CDF) plots, etc.
Dataset
The ISAdetect dataset was used to create the i386, AMD64, MIPSEL, MIPS64EL, ARM64, ARMEL, PowerPC, PPC64, and SH4 reference distributions.
Todo
- Adding the ability to use OPTICS for automatic clustering. It would be nice to automate the entire workflow, going straight from an input file to data type identification. Currently this is not possible because
epsandmin_samplesneed to be adjusted manually in order ensure meaningful results when using DBSCAN. - Improving the UTF-8 english data reference distribution. Rather than derive it from text extracted from an ebook, samples should be drawn from hard-coded text data in executable binaries.
- Creating reference distributions for AVR and Xtensa
- update the code with docstrings and comments
